Ophthalmologe (2022)
Julian Wolf, Thabo Lapp, Thomas Reinhard, Hansjürgen Agostini, Günther Schlunck, Clemens Lange
Number of citations (crossref.org): Loading....

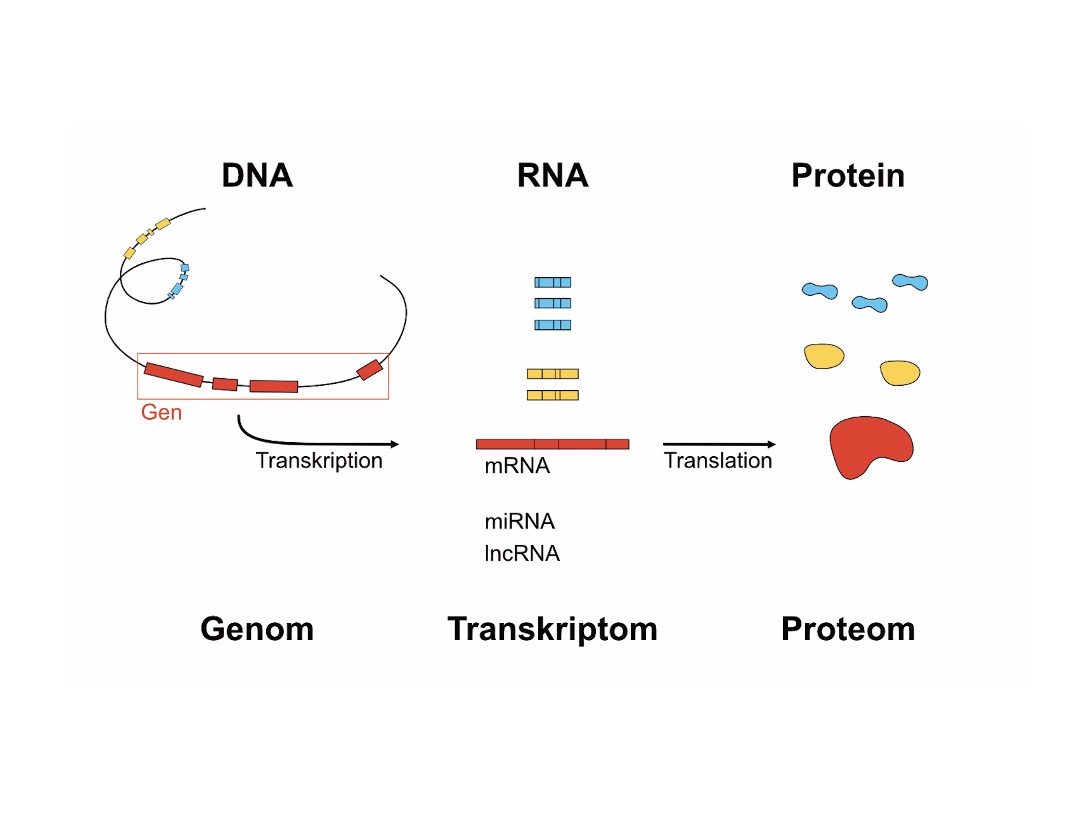
Background: Gene expression analysis using RNA sequencing has helped
to improve the understanding of many diseases. Databases, such as the Gene Expression Omnibus
database of the National Center for Biotechnology Information provide RNA sequencing raw data
from various diseased tissue types but their analysis requires advanced bioinformatics skills.
Therefore, specific ocular databases provide the transcriptional profiles of different ocular
tissues and in addition enable intuitive web-based data analysis.
Objective: The aim of this narrative review is to provide an overview
of ocular transcriptome databases and to compare them with the Human Eye Transcriptome
Atlas newly established in Freiburg.
Methods: PubMed literature search.
Results: A total of nine ocular transcriptome databases focusing on different
aspects were identified. The iSyTE and Express platforms specialize in gene expression during
lens and retinal development in mice, whereas retina.tigem.it, Eye in a Disk, and Spectacle
focus on selected ocular tissues such as the retina. Spectacle, UCSC Cell Browser and Single
Cell Portal allow intuitive exploration of single cell RNA sequencing data derived from retinal,
choroid, cornea, iris, trabecular meshwork and sclera specimens. The microarray profiles of a
variety of healthy ocular tissues are included in the Ocular Tissue Database. The Human Eye
Transcriptome Atlas provides the largest collection of different ocular tissue types, contains
the highest number of ocular diseases and is characterized by a high level of quality achieved
by methodological consistency.
Conclusion: Ocular transcriptome databases provide comprehensive and intuitive insights
into the transcriptional profiles of a variety of healthy and diseased ocular tissues. Thus, they
improve our understanding of the underlying molecular mediators, support hypothesis generation and
help in the search for new diagnostic and therapeutic targets for various ocular diseases.