Scientific Reports (2020)
Stefaniya Boneva, Anja Schlecht, Peipei Zhang, Daniel Böhringer, Thabo Lapp, Hans Mittelviefhaus, Thomas Reinhard, Claudia Auw-Haedrich, Günther Schlunck, Julian Wolf, Clemens Lange
Number of citations (crossref.org): Loading....

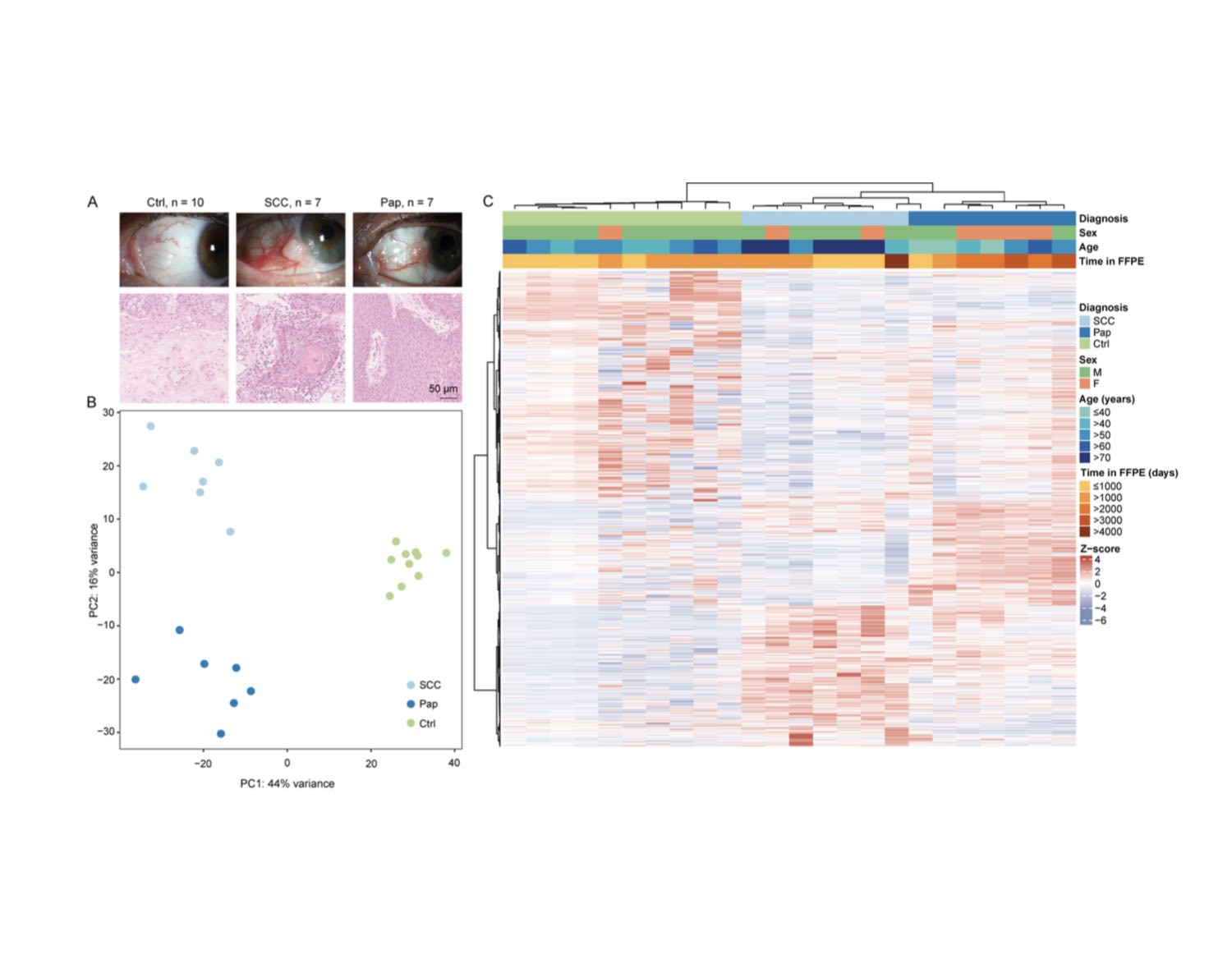
Recent advances in the field of biomedical research allow for elucidation of the transcriptional signature of rare tumors such as conjunctival squamous cell carcinoma (SCC). In this study we compare its expression profile to conjunctival papilloma (Pap) and healthy conjunctival tissue (Ctrl) and develop a classification tool to differentiate these entities. Seven conjunctival SCC, seven Pap and ten Ctrl were formalin-fixed and paraffin-embedded (FFPE) and analyzed using Massive Analysis of cDNA Ends (MACE) RNA sequencing. Differentially expressed genes (DEG) and gene ontology (GO) clusters were explored and the abundance of involved cell types was quantified by xCell. Finally, a classification model was developed to distinguish SCC from Pap and Ctrl. Among the most prominent DEG in SCC a plethora of keratins were upregulated when compared to Pap and Ctrl. xCell analysis revealed an enrichment of immune cells, including activated dendritic cells and T-helper type 1 cells (Th1), in SCC when compared to Ctrl. The generated classification model could reliably discriminate between the three entities according to the expression pattern of 30 factors. This study provides a transcriptome-wide gene expression profile of rare conjunctival SCC. The analysis identifies distinct keratins, as well as dendritic and Th1 cells as important mediators in SCC. Finally, the provided gene expression classifier may become an aid to the conventional histological classification of conjunctival tumors in uncertain cases.